Dask performance benchmarking put to the test: Fixing a pandas bottleneck
Getting notified of a significant performance regression the day before release sucks, but quickly identifying and resolving it feels great!
We were getting set up at our booth at JupyterCon 2023 when we received a notification: An engineer on our team had spotted a significant performance regression in Dask. With an impact of 40% increased runtime, it blocked the release planned for the next day!
Luckily, the other attendees still focused on coffee and breakfast, so we commandeered an abandoned table next to our booth and got to work.
Performance testing at Coiled
The performance problem had been flagged by the automated performance testing for Dask that we developed at Coiled.
If you have not read Guido Imperiale's blog post on our approach to performance testing, here is a summary:
With coiled/benchmarks, we created a benchmark suite that contains a variety of common workloads and operations with Dask, including standardized ones like the h2oai/db-benchmark.
It also contains tooling that allows us to do two things:
- Automatically detect performance regressions in Dask and raise them as issues.
- Run A/B tests to assess the performance impact of different versions of Dask, upstream packages, or cluster configurations.
While the former started this journey, the latter will also come in handy soon.
Identifying the problem
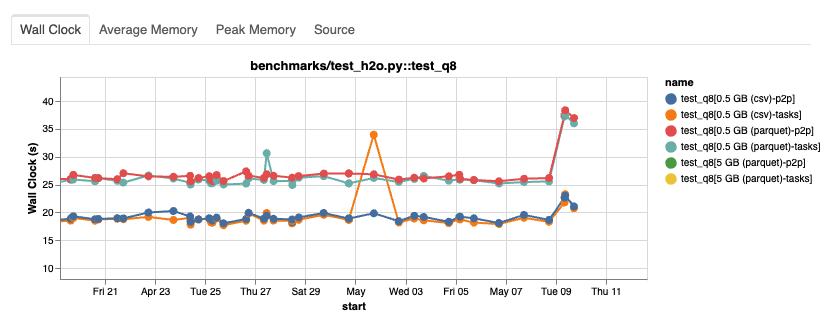
Our automated regression testing had alerted us that test_h2o.py::test_q8 had experienced a significant increase in runtime across all data sizes and file formats.
From the historical report of our benchmarking suite, we could see that dask/dask and dask/distributed were unlikely to be the culprit:
Nothing had changed on dask/dask when the performance started to degrade, and there was only one unrelated change on dask/distributed.
That left us with the Coiled platform and upstream packages as possible candidates.
After digging deeper into the cluster data, we noticed that pandas had been upgraded from 1.5.3 to 2.0.1.
A major upgrade to pandas at the same time a dataframe-based workload shows degrading performance? That's suspicious!
To confirm this suspicion, we ran an A/B test based on the current Dask release (2023.4.1 at the time), testing the impact of the pandas upgrade.
The results were clear: The runtime increased significantly with pandas=2.0.1 (sample).
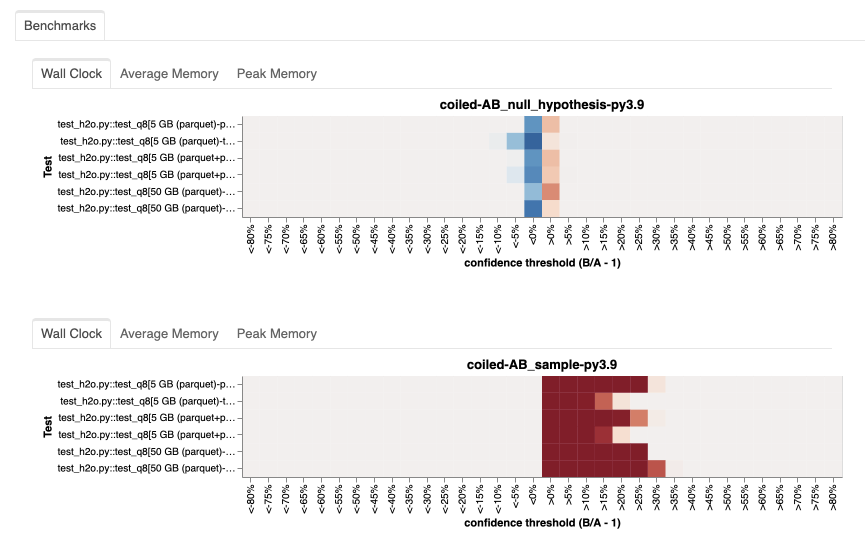
Having shown that pandas caused for the performance degradation and that we could reproduce it with the current Dask release, our release process for 2023.5.0 was cleared.
To further analyze the problem, we also derived a minimal local reproducer from the original workload:
from dask.distributed import Client
client = Client()
uri = "s3://coiled-datasets/h2o-benchmark/N_1e7_K_1e2_parquet/*.parquet"
ddf = dd.read_parquet(uri, engine="pyarrow", storage_options={"anon": True}).persist()
wait(ddf)
ddf = ddf[["id6", "v1", "v2", "v3"]]
(
ddf[~ddf["v3"].isna()][["id6", "v3"]]
.groupby("id6", dropna=False, observed=True)
.apply(
lambda x: x.nlargest(2, columns="v3"),
meta={"id6": "Int64", "v3": "float64"},
)[["v3"]]
).compute()
Investigating the pandas performance degradation
The only user-visible thing that changed between pandas 1.5.3 and pandas 2.0.1 was the default value
of group_keys in GroupBy. Switching to group_keys=False with version 2.0.1
got us back to the initial runtime.
Now that we knew that pandas was to blame for the performance degradation, we had to create a
reproducer in plain pandas to help fix the issue.
df = pd.DataFrame(
{
"foo": np.random.randint(1, 50_000, (100_000, )),
"bar": np.random.randint(1, 100_000, (100_000, )),
},
)
df.groupby(
"foo", group_keys=False
).apply(lambda x: x.nlargest(2, columns="bar"))
group_keys=False: approx. 11 secondsgroup_keys=True: approx. 15 seconds
Experimenting a bit showed us that the bottleneck got even worse while increasing the number of
groups during the groupby calculation. We settled on this version which is 30% slower with
group_keys=True, enough to be able to troubleshoot the problem. There was no obvious reason
why the changed value should bring a significant slowdown.
%prun showed us that the time was almost exclusively spent in a post-processing step that
combines all groups via concat.
Addressing the performance degradation
Let's look at how both cases differ. The new version passes the grouping levels to concat, which
are used to construct the resulting Index levels. This shouldn't be that slow though. Investigations
showed that this runs through a code-path that is very inefficient!
Looking closer at the results of %prun pointed us to one specific loop that took up most of
the runtime. This loop calculates the codes for the resulting index based on the provided
levels. It's slow, really slow! Every single element provided as keys, which
represent the number of groups, is checked against the whole level, which explains our previous
observation that the runtime got worse with an increasing number of groups. You can check out the
pandas user guide
if you are interested in situations where this is useful. Fortunately, we have a convenient
advantage in case of groupby. We know beforehand that every key equals the specific level.
We added a fast-path that exploits this knowledge getting the runtime of this step more or less to
zero.
This change resulted in a small PR that cut
the runtime of group_keys=True to approximately 11 seconds as well.
Conclusion
Now that we made our pandas reproducer run 30% faster, we have to check whether we accomplished our initial objective. Re-running the local Dask reproducer should give us an idea about the impact on Dask. We got the performance down to 22 seconds as well! Promising news that saved our plans for the evening!
Unfortunately, we had to wait until pandas 2.0.2 was released to run a proper benchmark.
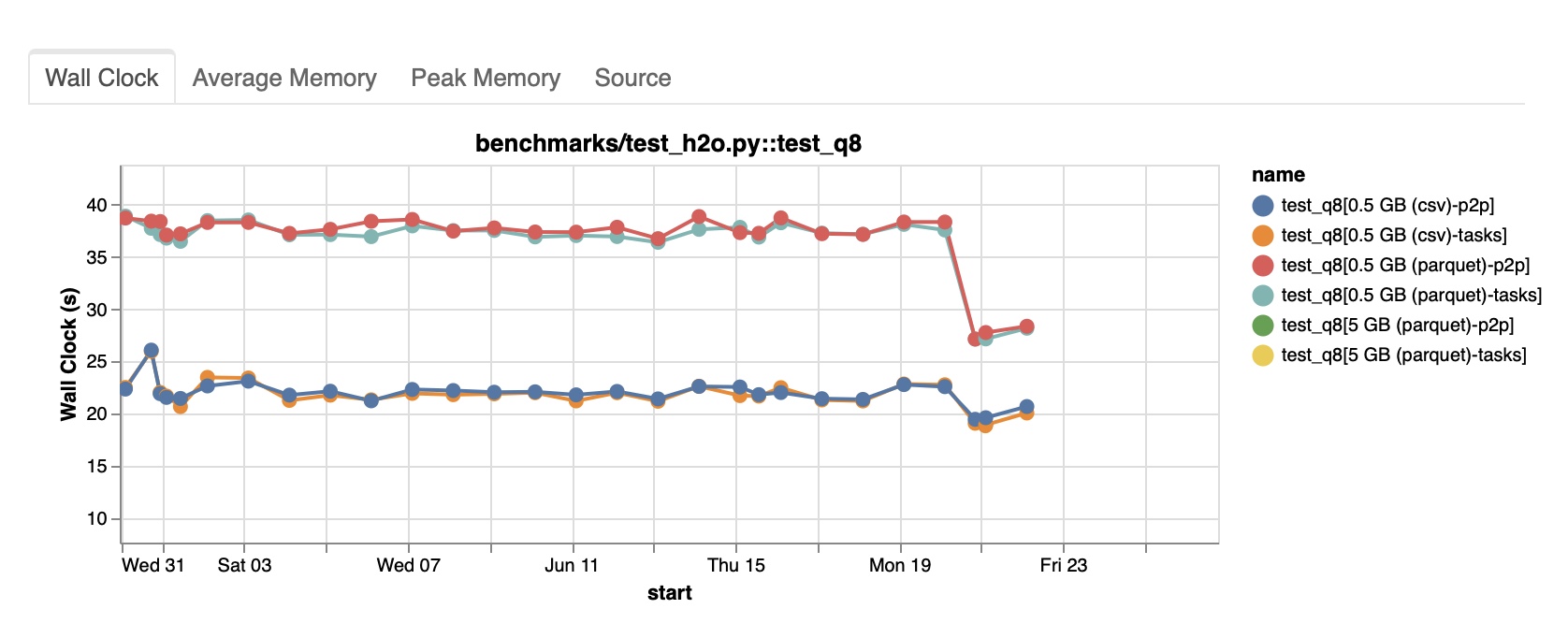
This looks great! Our small pandas change translated to our Dask query and got performance back to the previous level!